 It was a delight today to chat with Monica Coenraads, Executive Director of the Rett Syndrome Research Trust. The RSRT has teamed up with a deeply focused world-class team of research scientists to translate the fruits of basic research on Rett syndrome into viable cures. Whether you are a scientist, student or concerned family member, you will learn a lot from exploring the RSRT website, blog as well as this short video lecture. Just by a strange, unanticipated coincidence, today marks the 10-year annivesary of the identification of MeCP2 as the underlying gene for Rett syndrome. Click here for prior blog posts on Rett syndrome. (click here for podcast)
It was a delight today to chat with Monica Coenraads, Executive Director of the Rett Syndrome Research Trust. The RSRT has teamed up with a deeply focused world-class team of research scientists to translate the fruits of basic research on Rett syndrome into viable cures. Whether you are a scientist, student or concerned family member, you will learn a lot from exploring the RSRT website, blog as well as this short video lecture. Just by a strange, unanticipated coincidence, today marks the 10-year annivesary of the identification of MeCP2 as the underlying gene for Rett syndrome. Click here for prior blog posts on Rett syndrome. (click here for podcast)
Posts Tagged ‘Development’
podcast: Rett Syndrome Research Trust
Posted in MECP2, tagged Development, economics, Epigenetics, Genetic Disorders, Genetic testing, Mental disorder, Mental health, Podcast, Rett Syndrome on October 1, 2009| Leave a Comment »
Too much yin and not enough yang in cortical networks of MeCP2 mutant mice
Posted in MECP2, tagged autism, Development, Epigenetics, Gene, Gene expression, Long-Term Potentiation, MECP2, Mental disorder, Mental health, Mental retardation, Neural network, Neuron, Rett Syndrome on September 30, 2009| 1 Comment »
- Image via Wikipedia
In previous posts, we have explored some of the basic molecular (de-repression of chromatin structure) and cellular (excess synaptogenesis) consequences of mutations in the MeCP2 gene – a.k.a the gene whose loss of function gives rise to Rett syndrome. One of the more difficult aspects of understanding how a mutation in a lowly gene can give rise to changes in cognitive function is bridging a conceptual gap between biochemical functions of a gene product — to its effects on neural network structure and dynamics. Sure, we can readily acknowledge that neural computations underlie our mental life and that these neurons are simply cells that link-up in special ways – but just what is it about the “connecting up part” that goes wrong during developmental disorders?
In a recent paper entitled, “Intact Long-Term Potentiation but Reduced Connectivity between Neocortical Layer 5 Pyramidal Neurons in a Mouse Model of Rett Syndrome” [doi: 10.1523/jneurosci.1019-09.2009] Vardhan Dani and Sacha Nelson explore this question in great detail. They address the question by directly measuring the strength of neural connections between pyramidal cells in the somatosensory cortex of healthy and MeCP2 mutant mice. In earlier reports, MeCP2 neurons showed weaker neurotransmission and weaker plasticity (an ability to change the strength of interconnection – often estimated by a property known as “long term potentiation” (LTP – see video)). In this paper, the authors examined the connectivity of cortical cells using an electrophysiological method known as patch clamp recording and found that early in development, the LTP induction was comparable in healthy and MeCP2 mutant animals, and even so once the animals were old enough to show cognitive symptoms. During these early stages of development, there were also no differences between baseline neurotransmission between cortical cells in normal and MeCP2 mice. Hmmm – no differences? Yes, during the early stages of development, there were no differences between genetic groups – however – once the team examined later stages of development (4 weeks of age) it was apparent that the MeCP2 animals had weaker amplitudes of cortical-cortical excitatory neurotransmission. Closer comparisons of when the baseline and LTP deficits occurred, suggested that the LTP deficits are secondary to baseline strength of neurotransmission and connectivity in the developing cortex in MeCP2 animals.
So it seems that MeCP2 can alter the excitatory connection strength of cortical cells. In the discussion of the paper, the authors point out the importance of a proper balance of inhibition and excitation (yin and yang, if you will) in the construction or “connecting up part” of neural networks. Just as Rett syndrome may arise due to such a problem in the proper linking-up of cells – who use their excitatory and inhibitory connections to establish balanced feedback loops – so too may other developmental disorders such as autism, Down’s syndrome, fragile X-linked mental retardation arise from an improper balance of inhibition and excitation.
Getting to know my inner shark
Posted in Uncategorized, tagged Animal, Biology, Development, evolution, Health, Human, Human body, Mammal, Neil Shubin, Your Inner Fish: A Journey into the 3.5-Billion-Year History of the Human Body on September 29, 2009| Leave a Comment »
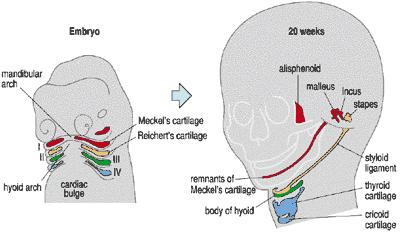 Am having a wonderful time reading, “Your Inner Fish” by Professor Neil Shubin – an exploration into the deep evolutionary roots of the human body. Amazed to contemplate the embryonic structures known as the branchial arches, or gill arches – which we share with sharks! – and the role of the gcm2 gene that is expressed in these arches and controls salt balance in humans and fish. Pharyngula has a wonderful post on this !!
Am having a wonderful time reading, “Your Inner Fish” by Professor Neil Shubin – an exploration into the deep evolutionary roots of the human body. Amazed to contemplate the embryonic structures known as the branchial arches, or gill arches – which we share with sharks! – and the role of the gcm2 gene that is expressed in these arches and controls salt balance in humans and fish. Pharyngula has a wonderful post on this !!
Hoping to find more deep evolutionary roots of mind and brain.

Support staff deserves some of the blame for Rett syndrome deficits
Posted in MECP2, White matter, tagged Ari Gold, autism, Development, Glial cell, MECP2, Mental disorder, Neural development, Neuron, Rett Syndrome, White matter on September 28, 2009| Leave a Comment »
 Celebrities and politicians are known for their love of the spotlight. “Me, me, me!” are the words to get ahead by in our modern media circus. As well, it can even be – in the unglamorous world of science – where, in characteristically geeky form, the conventional wisdom is to shout, “my hypothesis, my hypothesis, my hypothesis!”. Once, for example, I had a grad school professor say she was not allowed by her department to teach about glial cells in her brain development class. Another distinguished professor once told me, “don’t even bother sending a grant in, if it is focused on white matter“. No sir, it appears that modern neuroscience shall only focus on one main hypothesis – the neuron doctrine and not on the lowly support cells (astrocytes, oligodendrocytes & microglia) that, actually, make up more than 90% of the human brain. Hmmm, who would have thought to find such a cult of neuronal celebrity in the halls of academia?
Celebrities and politicians are known for their love of the spotlight. “Me, me, me!” are the words to get ahead by in our modern media circus. As well, it can even be – in the unglamorous world of science – where, in characteristically geeky form, the conventional wisdom is to shout, “my hypothesis, my hypothesis, my hypothesis!”. Once, for example, I had a grad school professor say she was not allowed by her department to teach about glial cells in her brain development class. Another distinguished professor once told me, “don’t even bother sending a grant in, if it is focused on white matter“. No sir, it appears that modern neuroscience shall only focus on one main hypothesis – the neuron doctrine and not on the lowly support cells (astrocytes, oligodendrocytes & microglia) that, actually, make up more than 90% of the human brain. Hmmm, who would have thought to find such a cult of neuronal celebrity in the halls of academia?
With this in mind, I really enjoyed the recent paper “Rett Syndrome Astrocytes Are Abnormal and Spread MeCP2 Deficiency through Gap Junctions” [doi:10.1523/jneurosci.0324-09.2009] by Maezawa and colleagues. The authors point out several critical gaps in the literature – namely that the expression of MeCP2 (the gene that, when mutated, gives rise to Rett syndrome) in neurons does NOT account for all of the many facets of the syndrome. For example, when MeCP2 is deleted only in neurons (in a mouse model), it results in a milder form of abnormal neural development than when deleted in all CNS cell types ( the full mouse syndrome: stereotypic forelimb motions, tremor, motor and social behavioral abnormalities, seizures, hypoactivity, anxiety-like behavior and learning/memory deficits). Also, it is not possible to reverse or rescue these deficits when a functional version of MeCP2 is expressed under a neuron-specific promoter. However, when re-expressed under its endogenous promoter – it is possible to rescue the syndrome (free access article).
The authors thus looked much more closely at the expression of MeCP2 and found that they could indeed visualize the expression of the MeCP2 protein in cultured ASTROCYTES – who are a very, very important type of support cell (just think of the personal secretary Lloyd to Ari Gold on the TV show “Entourage”). The team then examined how astrocytes that lack 80% of the expression of MeCP2 might interact with neurons – the very cells they normally support with secretions of growth factors and cytokines. It turns out that both normal and MeCP2-deficient neurons do not thrive when co-cultured with astrocytes that have weak MeCP2 expression. The team reports that dendritic length is reduced after a day and also a fews days of co-culture, suggesting that the MeCP2-deficient astrocytes are failing to provide the proper trophic support for their neuronal celebrity counterparts. Short dendrites are generally considered a bad-thing since this would predict poorer connectivity, and poorer cognition across the brain.
Hence, it seems that the lowly astrocyte is far more important in understanding what goes wrong in Rett syndrome. Ironically, in this case however, the celebrity status of the neuron remains untarnished as astrocytes can now be blamed for the consequences of MeCP2 mutations. The authors suggest that treatment of Rett syndrome via astrocytes is a worthwhile avenue of investigation. This new direction in the search for a cure will be an exciting story to follow!
resourceblog: Understanding the molecular basis of cognitive and social impairment in the autism spectrum disorders
Posted in HDACs, MECP2, tagged autism, Autism spectrum, Development, DNA, DNA methylation, Epigenetics, Gene, Gene expression, HDAC, Mental disorder, Mental health, Mutation, Rett Syndrome on September 24, 2009| Leave a Comment »

- Image via Wikipedia
The cognitive and emotional impairments in the autism spectrum disorders can be difficult for parents and siblings to understand and cope with. Here are some graphics and videos that might assist in understanding how genetic mutations and epigenetic modifications can lead to various forms of social withdrawl commonly observed in the autism spectrum disorders in children.
In this post, the focus is just on the MecP2 gene – where mutations are known to give rise to Rett Syndrome – one of the autism spectrum disorders. I’ll try and lay out some of the key steps in the typical bare-bones-link-infested-blogger-fashion – starting with mutations in the MecP2 gene. Disclaimer: there are several fuzzy areas and leaps of faith in the points and mouse model evidence below, and there are many other genes associated with various aspects of autism spectrum disorders that may or may not work in this fashion. Nevertheless, still it seems one can begin to pull a mechanistic thread from gene to social behavior Stay tuned for more on this topic.
1. The MecP2 gene encodes a protein that binds to 5-Methylcytosine – very simply – a regular cytosine reside with an extra methyl group added at position 5. Look at the extra -CH3 group on the cytosine residue in the picture at right. See? That’s a 5-methylcyctosine residue – and it pairs in the DNA double helix with guanosine (G) in the same fashion as does the regular cyctosine reside (C).  OK, now, mutations in the gene that encode the MecP2 gene – such as those found at Arginine residue 133 and Serine residue 134 impair the ability of the protein to bind to these 5-Methylcyctosine residues.
OK, now, mutations in the gene that encode the MecP2 gene – such as those found at Arginine residue 133 and Serine residue 134 impair the ability of the protein to bind to these 5-Methylcyctosine residues.  The figure at left illustrates this, and shows how the MecP2 protein lines up with the bulky yellow 5-Methylcytosine residues in the blue DNA double helix during binding.
The figure at left illustrates this, and shows how the MecP2 protein lines up with the bulky yellow 5-Methylcytosine residues in the blue DNA double helix during binding.
2. When the MecP2 protein is bound to the methylated DNA, it serves as a binding site for another type of protein – an HDAC or histone deacetylase. The binding of MecP2 and HDAC (and other proteins (see p172 section 5.3 of this online book “Chromatin Structure and Gene Expression“)). The binding of the eponymously named HDAC’s leads to the “de-acetylation” of proteins known as histones. The movie below illustrates how histone “de-acetylation” leads to the condensation of DNA structure and repression or shutting down of gene expression (when the DNA is tightly coiled, it is inaccessible to transcription factors). Hence: DNA methylation leads (via MecP2, HDAC binding) to a repression on gene expression.
3. When mutated forms of MecP2 cannot bind, the net result is MORE acetylation and MORE gene expression. As covered previously here, this may not be a good thing during brain development since more gene expression can induce the formation of more synapses and – possibly – lead to neural networks that fail to grow and mature in the “normal” fashion. The figure at right  suggests that neural networks with too many synapses may not be appropriately connected and may be locked-in to sub-optimal architectures. Evidence for excessive synaptogenesis is abundant within the autism spectrum disorders. Neuroligins – a class of genes that have been implicated in autism are known to function in cell & synaptic adhesion (open access review here), and can alter the balance of excitation/inhibition when mutated – which seems consistent with this heuristic model of neural networks that can be too adhesive or sticky.
suggests that neural networks with too many synapses may not be appropriately connected and may be locked-in to sub-optimal architectures. Evidence for excessive synaptogenesis is abundant within the autism spectrum disorders. Neuroligins – a class of genes that have been implicated in autism are known to function in cell & synaptic adhesion (open access review here), and can alter the balance of excitation/inhibition when mutated – which seems consistent with this heuristic model of neural networks that can be too adhesive or sticky.
4. Cognitive and social impairment can result from poor-functioning neural networks containing, but not limited to the amygdala. The normal development of neural networks containing the forntal cortex and amygdala are important for proper social and emotional function. The last piece of the puzzle then would be to find evidence for developmental abnormalities in these networks and to show that such abnormalities mediate social and/or emotional function. Such evidence is abundant.
Regarding the effects of MecP2 however, we can consider the work of Adachi et al., who were able to delete the MecP2 gene – just in the amygdala – of (albeit, an adult) mouse. Doing so, led to the disruption of various emotional behaviors – BUT NOT – of various social interaction deficits that are observed when MecP2 is deleted in the entire forebrain. This was the case also when the team infused HDAC inhibitors into the amygdala suggesting that loss of transcriptional repression in the adult amygdala may underlie the emotional impariments seen in some autism spectrum disorders. Hence, such emotional impairments (anxiety etc.) might be treatable in adults (more on this result later and its implications for gene-therapy).
Whew! Admittedly, the more you know – the more you don’t know. True here, but still amazing to see the literature starting to interlink across human-genetic, mouse-genetic, human-functional-imaging levels of analysis. Hoping this rambling was helpful.
Epigenetic puppetmasters pull strings of cognitive development from a safe distance
Posted in HDACs, tagged autism, Biology, Development, DNA, Epigenetics, Gene, Gene expression, Genetics, Mental disorder, Mouse, Natural selection, Neural network, Rett Syndrome on September 21, 2009| 2 Comments »

- Image by eugene via Flickr
The homunculus (argument) is a pesky problem in cognitive science – a little guy who might suddenly appear when you propose a mechanism for decision making, spontaneous action or forethought etc. – and would take credit for the origination of the neural impulse. While there are many mechanistic models of decision making that have slain the little bugger – by invoking competition between past experience and memory as the source of new thoughts and ideas – one must always tread lightly, I suppose, to be wary that cognitive mechanisms are based completely in neural properties devoid of a homuncular source.
Still, the human mind must begin somewhere. After all, its just a ball of cells initially, and then a tube and then some more folds, layers, neurogenesis and neural migration etc. before maturing – miraculously – into a child that one day looks at you and says, “momma” or “dada”. How do these neural networks come into being? Who or what guides their development toward that unforgettable, “momma (dada)” moment? A somewhat homuncluar “genetic program” – whose instructions we can attribute to millions of years of natural selection? Did early hominid babies say “momma (dada)? Hmmm. Seems like we might be placing a lot of faith in the so-called “instructions” provided by the genome, but who am I to quibble.
On the other hand, you might find that the recent paper by Akhtar et al., “Histone Deacetylases 1 and 2 Form a Developmental Switch That Controls Excitatory Synapse Maturation and Function” [doi:10.1523/jneurosci.0097-09.2009] may change the way you think about cognitive development. The team explores the function of two very important epigenetic regulators of gene expression – histone deacetylases 1,2 (HDAC1, HDAC2) on the functionality of synapses in early developing mice and mature animals. By epigenetic, I refer to the role of these genes in regulating chromatin structure and not via direct, site-specific DNA binding. The way the HDAC genes work is by de-acetylating – removing acetyl groups – thus removing a electrostatic repulsion of acetyl groups (negative charge) on histone proteins with the phosphate backbone of DNA (also a negative charge). When the histone proteins carry such an acetyl group, they do NOT bind well to DNA (negative-negative charge repulsion) and the DNA molecule is more open and exposed to binding of transcription factors that activate gene expression. Thus if one (as Akhtar do) turns off a de-acetylating HDAC gene, then the resulting animal has a genome that is more open and exposed to transcription factor binding and gene expression. Less HDAC = more gene expression!
What were the effects on synaptic function? To summarize, the team found that in early development (neonatal mouse hippocampal cells) cells where the HDAC1 or 2 genes were turned off (either through pharmacologic blockers or via partial deletion of the gene(s) via lentivirus introduction of Cre recombinase) had more synapses and more synaptic electrical activity than did hippocampal cells from control animals. Keep in mind that the HDACs are located in the nucleus of the neuron and the synapses are far, far away. Amazingly – they are under the control of an epigenetic regulator of gene expression; hence, ahem, “epigenetic puppetmasters”. In adult cells, the knockdown of HDACs did not show the same effects on synaptic formation and activity. Rather the cells where HDAC2 was shut down showed less synaptic formation and activity (HDAC1 had no effect). Again, it is amazing to see effects on synaptic function regulated at vast distances. Neat!
The authors suggest that the epigenetic regulatory system of HDAC1 & 2 can serve to regulate the overall levels of synaptic formation during early cognitive development. If I understand their comments in the discussion, this may be because, you don’t necessarily want to have too many active synapses during the formation of a neural network. Might such networks might be prone to excitotoxic damage or perhaps to being locked-in to inefficient circuits? The authors note that HDACs interact with MecP2, a gene associated with Rett Syndrome – a developmental disorder (in many ways similar to autism) where neural networks underlying cognitive development in children fail to progress to support higher, more flexible forms of cognition. Surely the results of Akhtar et al., must be a key to understanding and treating these disorders.
Interestingly, here, the controller of these developmental phenotypes is not a “genetic program” but rather an epigenetic one, whose effects are wide-spread across the genome and heavily influenced by the environment. So no need for an homunculus here.
Animal model reveals timely insights into immuno-genetic risk of schizophrenia
Posted in Frontal cortex, Hippocampus, MHC loci, tagged Development, Epigenetics, Frontal lobe, schizophrenia on August 19, 2009| 1 Comment »

- Image via Wikipedia
Among the various (and few) significant results of recent landmark whole-genome analyses (involving more than 54,000 participants) on schizophrenia (covered here and here), there was really just one consistent result – linkage to the 6p21-22 region containing the immunological MHC loci. While there has been some despair among professional gene hunters, one man’s exasperation can sometimes be a source of great interest and opportunity for others – who – for many years – have suspected that early immunological infection was a key risk factor in the development of the disorder.
Such is the case in the recent paper, “Prenatal immune challenge induces developmental changes in the morphology of pyramidal neurons of the prefrontal cortex and hippocampus in rats” by Baharnoori et al., [doi: 10.1016/j.schres.2008.10.003]. In this paper, the authors point out that Emil Kraepelin, who first described the disorder we now call schizophrenia, had suggested that childhood inflammation of the head might be an important risk factor. Thus, the immunopathological hypothesis has been around since day 0 – a long time coming I suppose.
In their research article, Baharnoori and colleagues have taken this hypothesis and asked, in a straightforward way, what the consequences of an immunological challenge on the developing brain might look like. To evaluate this question, the team used a Sprague-Dawley rat model and injected pregnant females (intraperitoneally on embryonic day 16) with a substance known as lipopolysaccharide (LPS) which is known to mimic an infection and initiate an immune response (in a manner that would normally depend on the MHC loci found on 6p21-22). Once the injections were made, the team was then able to assess the consequences to various aspects of brain and behavior.
In this paper, the team focused their analysis on the development of the frontal cortex and the hippocampus – 2 regions that are known to function poorly in schizophrenia. They used a very, very focused probe of development – namely the overall shape, branching structure and spine formations on pyramidal cells in these regions – via a method known as Golgi-Cox staining. The team presents a series of fantastically detailed images of single pyramidal cells (taken from postnatal day 10, 35 and 60) from animals who’s mothers were immunologically challenged and those who were unexposed to LPS.
Briefly, the team finds that the prenatal exposure to LPS had the effect of reducing the number of dendritic spines (these are the aspects of a neuron that are used to make synaptic connections with other neurons) in the developing offspring. Other aspects of neuronal shape were also affected in the treated animals – basically amounting to a less branchy, less spiny – less connectable – neuron. If that’s not a basis for a cognitive disorder than what else is? Indeed, the authors point out that such spines are targets – in early development – for interneurons that are essential for long-range gamma oscillations that help distant brain regions function together in a coherent manner (something that notably does not happen in schizophrenia).
Thus, there is many a reason (54,000 strong) to want to better understand the neuro-immuno-genetic-developmental mechanisms that can alter neuronal structure. Exciting progress in the face of recent genetic setbacks!

Transcriptional co-expression yields early clues to genetic influences on neural network structure/function (computation)
Posted in RGS4, tagged Development, Gene expression on August 18, 2009| Leave a Comment »

- Image via Wikipedia
In the lowly worm Caenorhabdritis elegans, it has been long possible to understand the exact lineage of each of its 959 somatic cells. That is, one can know for each and every cell, who its parental cell was, and grand-parental cell etc., back until the very first cell division (feast your eyes on these movies of C. elegans development). Similarly, it is possible to follow what networks of genes are transcribed (turned on/off) as these cellular divisions and differentiations occur. In this sense, one can reconstruct the transcriptional events occuring within the nucleus of a cell with outward changes in cell structure and migration as the lowly li’l worm develops.
If, for example, there were genes that led to abberant behavior in the worm, then it would be possible to query when and where such a gene was expressed and under who’s transcriptional control. Such a tool would be powerful and useful indeed – especially if there were genetically-based abberant behavioral disorders in worms. Nice to be a lowly worm (psychiatrist) these days.
So, what about human brains? and human genes that have been correlated with changes in brain structure, brain activity and/or brain disorders? Are there tools that allow us to reconstruct an outward cellular lineage and correlate it with a transcriptional lineage? Where might genes for abberant brain function lie in such a lineage? – perhaps early in the course of brain development, with many subsequent genes under its control and whose expression mediates the development of many daughter and granddaugter cells? Or perhaps mental illness risk genes have little regulatory oversight and have rather specific effects on a small number of specialized cells later in the course of development? Wouldn’t we like to know!
With this query in mind, it was fun to read a recent article entitled, “The organization of the transcriptional network in specific neuronal classes” by Winden et al., [free access doi: 10.1038/msb.2009.46]. This article describes an amazing bioinformatic open-public use tool called, “Weighted Gene Co-Expression Network Analysis” which seems to have been developed in the lab of Steve Horvath at UCLA (one of the co-authors). The authors examined the gene expression data from an array of 12 different adult neuronal cell types (this analysis was performed using mouse brain tissue) and used the WGCNA method to organize the patterns of gene expression and then asked how they relate to different cell morphologies and physiological attributes (such as firing patterns). In this way, they are beginning to construct a genetic road map of mammalian brain development that is much like that for C. elegans.
To me, the really exciting thing about this particular analysis (they also have used this method to compare gene expression in a brain-region-specific way in humans vs. chimpanzees) is that the different cell types in the brain perform different – here comes the punchline – computations! Thus, if there be a genetic code for neuron structure/function (and from the work of Horvath and colleagues, it looks like there be), then it should be possible to begin to assign these module-specific genetic factors to computations or computational properties of the brain – and have a more mechanistic synthesis of genetic influences on cognition.
A few examples from the paper include different transcriptional co-expression networks (modules) for glutamateric and GABAergic cell types, which have distinct functions in the regulation of neural dynamics (ie. computation). Further analysis yielded different co-expression modules for the development of different types of interneurons. The team also finds different co-expression modules for aspects of cell-firing and synaptic structure which would very likely have effects on neural-network dynamics. Also, there is an analysis of the RGS4 knockout mouse and a query into the specificity of the co-expression module that contains RGS4 (very few abberant gene expression changes outside the RGS4 module) which – since RGS4 has been associated with schizophrenia – reveals clues on the expected consequences of RGS4 mutations in humans.
There is really too much to cover in a single blog post, so am going to dig in to the data in more detail and report back later. An amazing tool graciously shared with the community!
White-matter correlates of gene penetrance reveal key brain circuits for dystonia
Posted in Cerebellum, Motor cortex, Thalamus, Thap1, Torsin A, tagged Biology, Development, Diffusion MRI, Disease, DNA, Dystonia, Gene, Genetics, Mental disorder, Mutation, White matter on August 14, 2009| Leave a Comment »

- Image via Wikipedia
Within the genetic news flow, there is often, and rightly so, much celebration when a gene for a disease is identified. This is indeed an important first step, but often, the slogging from that point to a treatment – and the many small breakthroughs along the way – can go unnoticed. One reason why these 2nd (3rd, 4th, 5th …) steps are so difficult, is that in some cases, folks who carry “the gene” variant for a particular disorder, do not, in fact, display symptoms of the disorder.
Huh? One can carry the risk variant – or many risk variants – and not show any signs of illness? Yes, this is an example of what geneticists refer to as variable penetrance, or the notion of carrying a mutation, but not outwardly displaying the mutant phenotype. This, is one of the main reasons why genes are not deterministic, but much more probablistic in their influence of human development.
Of course, in the brain, such complexities exist, perhaps even moreso. For example, take the neurological condition known as dystonia, a movement disorder that, according to the Dystonia Medical Research Foundation, “causes the muscles to contract and spasm involuntarily. The neurological mechanism that makes muscles relax when they are not in use does not function properly. Opposing muscles often contract simultaneously as if they are “competing” for control of a body part. The involuntary muscle contractions force the body into repetitive and often twisting movements as well as awkward, irregular postures.” Presently there are more than a dozen genes and/or chromosomal loci that are associated with dystonia – two of the major genes, DYT1 and DYT6 – having been identified as factors in early onset forms of dystonia. Now as we enter the era of personal genomes, an individual can assess their (own, child’s, preimplantion embryo’s!) genetic risk for such rare genetic variants – whose effects may not be visible until age 12 or older. In the case of DYT1, this rare mutation (a GAG deletion at position 946 which causes a loss of a glutamate residue in the torsin A protein) gives rise to dystonia in about 30-40% of carriers. So, how might these genes work and why do some individuals develop dystonia and others do not? Indeed, these are the complexities that await in the great expanse between gene identification and treatment.
An inspection of the molecular aspects of torsin A (DYT1) show that it is a member of the AAA family of adenosine triphosphatases and is related to the Clp protease/heat shock family of genes that help to properly fold poly-peptide chains as they are secreted from the endoplasmic reticulum of the cell – a sort-of handyman, general purpose gene (expressed in almost every tissue in the body) that sits on an assembly line and hammers away to help make sure that proteins have the right shape as they come off their assembly lines. Not much of a clue for dystonia – hmm. Similarly, the THAP domain containing, apoptosis associated protein 1 (THAP1) gene (a.k.a. DYT6) is also expressed widely in the body and seems to function as a DNA binding protein that regulates aspects of cell cycle progression and apoptosis. Also not much an obvious clue to dystonia – hmm, hmm. Perhaps you can now see why the identification of “the gene” – something worth celebrating – can just leave you aghast at how much more you don’t know.
That these genes influence an early developmental form of the disorder suggests a possible developmental role for these rather generic cogs in the cellular machinery. But where? how? & why an effect in some folks and not others? To these questions, comes an amazing analysis of DYT1 and DYT6 carriers in the article entitled, “Cerebellothalamocortical Connectivity Regulates Penetrance in Dystonia” by Argyelan and colleagues [doi: 10.1523/JNEUROSCI.2300-09.2009]. In this article, the research team uses a method called diffusion tensor imaging (sensitive to white matter density) to examine brain structure and function among individuals who carry the mutations but either DO or DO NOT manifest the symptoms. By looking at white matter tracts (super highways of neural traffic) throughout the brain the team was able to ask whether some tracts were different in the 2 groups (as well as a group of unaffectd, non-carriers). In this way, the team can begin to better understand the causal pathway between these run-of-the-mill genes (torsin A and thap1) and the complex pattern of muscle spasms that arise from their mutations.
To get right to the findings, the team has discovered that in one particular tract, a superhighway known as “cerebellar outflow pathway in the white matter of lobule VI, adjacent to the dentate nucleus” (not as quaint as Route 66) that those participants that DO manifest dystonia had less tract integrity and connectivity there compared to those that DO NOT manifest and healthy controls (who have the most connectivity there). Subsequent measures of resting-state blood flow confirmed that the disruptions in white matter tracts were correlated with cerebellar outflow to the thalamus and – more importantly – with activity in areas of the motor cortex. The correlations were such that individuals who DO manifest dystonia had greater activity in the motor cortex (this is what dystonia really comes down to — too much activity in the motor cortex).
Thus the team were able to query gene carriers using their imaging methods and zero-in on “where in the brain” these generic proteins exert a detrimental effect. This seems to me, to be a huge step forward in understanding how a run-of-the-mill gene can alter brain function in such a profound way. Now that they’ve found the likely circuit (is it the white matter per se or the neurons?), more focus can be applied to how this circuit develops – and can be repaired.
SNORD115 confirms autism risk in 15q11-13 duplication mouse model
Posted in 5HTT, SNORD115, SNRPN, UBE3A, tagged Angelman Syndrome, autism, Biology, Development, Epigenetics, Eukaryotic, Gene, Genetics, Genomic imprinting, Prader-Willi syndrome, RNA on July 21, 2009| 1 Comment »

- Image via Wikipedia
One way to organize the great and growing body of research into autism is via a sort-of ‘top-down’ vs. ‘bottom-up’ perspective. From the ‘top-down’ one can read observational research that carefully catalogs the many & varied social and cognitive attributes that are associated with autism. Often times, these behavioral studies are coupled with neurochemical or neuroimaging studies that test whether variation in such biomarkers is correlated with aspects of autism. In this manner, the research aims to dig down into the physiology and biochemistry of the developing brain to find out what is different and what differences might predict the onset of autistic traits. At the deepest biological level – the bedrock, so to speak – are a number of genetic variations that have been correlated with autism. These genetic variants permit another research strategy – a ‘bottom-up’ strategy that allows investigators to ask, “what goes wrong when we manipulate this genetic variant?” While proponents of each strategy are painfully aware of the limitations of their own strategy – oft on the barbed-end of commentary from the other side – it is especially exciting when the ‘top-down’ and ‘bottom-up’ methods find themselves meeting in the agreement in the middle.
So is the case with Nakatani et al., “Abnormal Behavior in a Chromosome- Engineered Mouse Model for Human 15q11-13 Duplication Seen in Autism” [doi: 10.1016/j.cell.2009.04.024] who created a mouse that carries a 6.3 megabase duplication of a region in the mouse that luckily happens to be remarkably conserved in terms of gene identity and order with the 15q11-13 region in humans – a region that, when duplicated, is found in about 5% of cases with autism. [click here for maps of mouse human synteny/homology on human chr15] Thus the team was able to engineer mice with the duplication and ask, “what goes wrong?” and “does it resemble autism in any kind of meaningful way (afterall these are mice we’re dealing with)?”
Well, the results are rather astounding to me. Most amazing is the expression of a small nucleoar RNA (snoRNA) – SNORD115 (mouse-HBII52) – that function in the nucleolus of the cell, and plays a role in the alternative splicing of exon Vb of the 5HT2C receptor. The team then found that the editing of 5HTR2C was altered in the duplication mice and also that Ca++ signalling was increased when the 5HTR2C receptors were stimulated in the duplication mice (compared to controls). Thus, a role for altered serotonin function – which has been a longstanding finding in the ‘topdown’ approach – was met midway and affirmed by this ‘bottom-up’ approach! Also included in the paper are descriptions of the abberant social behaviors of the mice via a 3-chambered social interaction test where duplication mice were rather indifferent to a stranger mouse (wild-type mice often will hang out with each other).
Amazing stuff!
Another twist to the story is the way in which the 15q11-13 region displays a phenomenon known as genomic-imprinting, whereby only the mother or the father’s portion of the chromosome is expressed. For example, the authors show that the mouse duplication is ‘maternally imprinted’ meaning that that pups do not express the copy of the duplication that comes from the mother (its expression is shut down via epigenetic mechanisms that involve – wait for it – snoRNAs!) so the effects that they report are only from mice who obtained the duplication from their fathers. So, if you by chance were wondering why its so tough to sort out the genetic basis of autism – here’s one reason why. On top of this, the 5HTR2C gene is located on the X-chromosome which complicates the story even more in terms of sorting out the inheritance of the disorder.
Further weird & wild is the fact that the UBE3A gene (paternally imprinted) and the genetic cause of Angelman Syndrome sits in this region – as does the SNRPN gene (maternally imprinted) which encodes a protein that influences alternative RNA splicing and also gives rise to Prader-Willi syndrome. Thus, this tiny region of the genome, which carries so-called “small” RNAs can influence a multitude of developmental disabilities. Certainly, a region of the genome that merits further study!!
From the insula, rs4606 pulls a common thread back through developmental space and time
Posted in Amygdala, Insula, RGS2, tagged 23andMe, Anxiety, Development on July 7, 2009| 1 Comment »

- Image by freeparking via Flickr
Back in the day, when the fam would get together at my parents’ house, I would enjoy shuffling through their box of old photos. Looking at childhood pictures of myself and relatives, it was natural to compare our adult selves to the old pictures and look for similarities – emotional expressions, gestures, etc. – that have carried on through the years and are (were) a part of who we are (became) today. It’s always amazing what you think you can see, and if you’re like me, you may be somewhat amazed by how much of your adult self was already in full swing as a child. The manner in which the developing brain confers such stability over time and over generations (now I see my own childhood traits in my son – yikes!) is of course a timeworn question among families and scientists alike.
That the genome would contribute to cross generational parent-child similarities in personality and temperament is fairly obvious, but not so apparent is how the genome interacts with the environment to exert an influence on psychological development. Along this line of inquiry, a research article entitled, “Influence of RGS2 on anxiety-related temperament, personality, and brain function” by Smoller and colleagues [free access] provides an amazing perspective – from a single gene. RGS2, eponymously named as a regulator of G-protein signaling, was first identified as a factor that regulates emotional behavior in mice [PMID] and subsequently as a risk factor for schizophrenia [PMID] as well as anxiety disorders in humans [PMID]. In the current study, the team examined the temperament of children (119 families), personality of adults (744 undergraduates) and brain activity in adults (55 participants) to ascertain whether the adult risk for anxiety conferred by RGS2 might be related to actions of the gene that occur much earlier in development – such as on the systems that regulate temperament in children. Specifically, they focused on behavioral inhibition in children (shy, avoidant, restrained in novel situations) and introversion in adults – as these traits have been associated with increased risk for anxiety disorders.
What is so interesting to me is that RGS2 (particularly the G allele of the 3’UTR SNP rs4606) was found to be associated with both childhood temperament and adult personality. Thus, an introverted adult who looks through an old photo album and sees themselves to have been a shy or inhibited child, may be experiencing – to a small degree – the effects of the RGS2 gene. The team suggests, via additional brain imaging-genetic studies, that RGS2 is of particular relevance to activity in circuits containing the insular cortex and amygdala – when subjects perform an emotional face matching task.
My own 23andme record does not contain the rs4606 SNP but does contain the data for rs1819741 where a T allele was significantly associated with introversion. Since I’m a C/T heterozygote, I guess I’ll have to look a bit harder at my old pictures to see signs of behavioral inhibition.
Autism genetic risk modulating the healthy social brain
Posted in Amygdala, AVPR1a, tagged autism, Development, Genetic testing, Mental disorder on June 20, 2009| 2 Comments »
 Comparisons of “healthy” vs. “disordered” genomes in psychiatry have not yet revealed sequence differences that can reliably predict the onset of mental disability. Rather, such disability seems to arise from as-yet-undetermined complex, probablistic interactions of genetic risk and environmental factors over the course of development. With this as the case, the demarcations between “healthy” and “disordered” may not be distinct, but rather fuzzy and hence unworthy of labels that give a false impression of being discrete states.
Comparisons of “healthy” vs. “disordered” genomes in psychiatry have not yet revealed sequence differences that can reliably predict the onset of mental disability. Rather, such disability seems to arise from as-yet-undetermined complex, probablistic interactions of genetic risk and environmental factors over the course of development. With this as the case, the demarcations between “healthy” and “disordered” may not be distinct, but rather fuzzy and hence unworthy of labels that give a false impression of being discrete states.
One recent paper that speaks to this issue is by Meyer-Lindenberg et al., “Genetic variants in AVPR1A linked to autism predict amygdala activation and personality traits in healthy humans” [doi: 10.1038/mp.2008.54]. Here, they explore genetic variation in the AVPR1A gene – a receptor for the pro-social neuropeptide vasopressin – and how it can modulate the activity of the amygdala when subjects view human faces (vs. a geometric shapes control condition). Since it is well known that the amygdala responds to a wide range of social and emotional stimuli and that activation of the amygdala can enhance or prevent the storage of such emotional or socially arousing events – and – that this process goes awry in autism and in subjects with amygdala damage (the picture above shows that patients with amygdala damage do not focus on the eyes of human faces) – the investigators have indeed focused-in on a key set of neural processes. They find the variation in the RS1 and RS3 polymorhphic sites in AVPR1a do indeed correlate with amygdala activity in healthy controls who were carefully screened for no history of mental disability. A great example of folks who carry the “healthy” label, but also the genetic risk and the neural correlates of autism.
Ungroomed granddaughters protest epigenetic marks on BDNF
Posted in BDNF, tagged Development, Epigenetics, evolution on May 8, 2009| Leave a Comment »

- Image via Wikipedia
Among mammalian species, moms can have it rough. THEY do the foraging and the child rearing usually without the help of dad who may or may not be prancing about defending his territory or doing who knows what. The biological systems that manage such a predicament for the female would, not surprisingly, be highly regulated and, I imagine, a major target of natural selection. For example, conflicts between what’s best for the offspring and what’s best for mom could drive the evolution of genetic and epigenetic mechanisms that counter-balance the tendency of moms to conserve resources and for offspring to use resources. One such epigenetic mechamism that has been implicated in parent-offspring conflict is so-called genomic imprinting – wherein certain epigenetic marks (methylation of C*pG’s in many cases) leads to the expression of genes a parent-of-origin-type manner (eg. the gene inherited from mom might be expressed while the gene inherited from dad would be transcriptionally repressed).
With this link between epigenetics and maternal investment in mind (and with Mother’s Day around the corner) I enjoyed the recent paper, “Lasting Epigenetic Influence of Early-Life Adversity on the BDNF Gene” by Roth and colleagues [doi: 10.1016/j.biopsych.2008.11.028] where they measured the relationship between BDNF expression and methylation as a function of maternal behavior (in stressful and non-stressful) conditions. Like many other neuronally-expressed genes, stress seems to lead to more methylation, which can – sometimes – interfere with transcription. In the Roth et al., paper, BDNF seems to show this pattern as well since BDNF was downregulated about 50% in the prefrontal cortex of rat pups who were reared under stressful conditions. Concomitant increases in methylation in the pups (which was blocked with methyltransferase inhibitors) was examined as a possible reason for the BDNF downregulation. Most interestingly, the female pups who were raised by stressed moms – were themselves lousy moms (demonstrated poor licking and grooming behavior) and gave birth to pups (granddaughters) who also bore similar epigenetic marks on BDNF.
Is this maternal-care/epigenetics phenomena related to parent-offspring conflict? Perhaps so, or perhaps just a spandrel or an unintended consequence of other levels of regulation. It will be fun to explore this further. Until then – be sure to thank your GRANDmother on Mother’s Day! – or not, if your are poorly groomed.
Growin’ up in da (rhesus monkey) ‘hood protects against MAOA ‘low’lifes
Posted in MAOA, Uncategorized, tagged Development, Gene-Environment on May 5, 2009| Leave a Comment »

- Image by Ginger Me via Flickr
As I and many other 23andMe participants begin to confront our genetic innards, we will likely ask whether any of the information is predictive. Can we expect to read-off our genomic information and say, “I have risk for this, this, and this, and so I’ll change my life to compensate ?” Certainly, in the area of mental health, there are genetic variants that confer bits of risk toward anxiety, depression, cognitive decline etc., but does the raw genomic information – alone – form a basis for diagnosis and proscriptive change? In most cases, NO. Rather, the genome is not unlike a plant seed, that will produce full leafy greens in rich soil, but merely a few buds in poor soil.
A great example of this can be seen in the recent paper, “What is an “Adverse” Environment? Interactions of Rearing Experiences and MAOA Genotype in Rhesus Monkeys” by Karere er al. [doi: 10.1016/j.biopsych.2008.11.004]. In this paper, they compared the emotional development of rhesus monkey infants (n=473) who carry different versions of an MAOA promoter polymorphism – so-called ‘low’ vs. ‘high’ transcriptional level alleles – and also who were reared in different social contexts. Some of the existing literature on MAOA-environment interactions suggests that abuse or neglect during childhood predisposes individuals who carry the ‘low’ allele (this allele leads to less MAOA protein and less catabolism of 5HT and DA). In this study, the environment was varied according to numbers of social companions and physical size of the neighborhood – (i) a field enclosure with up to 150 mixed adults & children, (ii) corncrib enclosure with 1 adult male, 2-5 females and various child playmates, (iii) mother-only small enclosure, and (iv) no-mother nursury rearing.
Which environment led to the emotional reactivity (anxiety, aggression etc.) that has been previously associated with the MAOA ‘low’ allele? Interestingly, it was not the wild & wooly ‘field enclosure’ where infants can interact in a rich, species-typical manner. Rather, it was the MAOA ‘low’ genotypic infants raised in the smaller groups who showed more signs of emotional reactivity, with cage-mother-only-rearing being the most extreme group. The authors note that this finding may alter our expectations about what type of environment is optimal vs. adverse and suggest that in the smaller enclosures, the relative isolation underlies the development of anxiety.
From a more general perspective, this study raises questions about how we – humans – should interpret our genomic information. What environmental conditions enhance or protect us from the potential genetic risk we carry? How did my early rearing interact with my MAOA allele? Something to discuss on Mother’s Day.
What’s a stubborn parent of a stubborn child to do? and other infinitely recurrent GxE interactions
Posted in DAT, Dopamine, tagged Development on April 29, 2009| Leave a Comment »

- Image via Wikipedia
The roles of nature and nurture in child development have never been easy to disentangle. Parents, in particular, seem to know this all too well, when it comes to their own children. For example, when one of my children throws a tantrum, my wife can be mercilessly quick to point out that “those are your genes at work “. I for one, can’t help but admire Mother Nature’s sense of justice (or is it humor?) as I’m forced to grapple with an unreconcilable 5-year old. What can I do? How can I get some type of optimal gene-by-environment (parenting style) going here? Afterall, they are MY genes (expressed in said unreconcilable 5-year old) right? Can I break out of the infinitely recurrent loop of me (my genes) trying to positively interact with my child (also my genes). What’s a stubborn parent of a stubborn child to do?
In thinking about this, it was great to read a recent article by Lee and colleagues entitled, “Association of maternal dopamine transporter genotype with negative parenting: evidence for gene x environment interaction with child disruptive behavior” [doi: 10.1038/mp.2008.102]. In this article, the team examined how children (4 to 7 years old) interacted with their mothers during a session where they were induced to cooperate in tasks involving free play with specific toys, tasks involving organizing items in a room and several pencil and paper tasks. A set of observations were made (through 1-way glass) on aspects of parenting (negative feedback or contact, positive feedback and encouragement, and, total number of maternal commands).
In principle, the complexities of whose genes & behavior is influencing whose in such a situation are vast. The authors point out that such interactions can be divided into passive GxE wherein children with certain genes (lets say genes for stubborness) may have inherited those genes from parents who exhibit a stubborn (negative) parenting style – hence leading to correlations in child genotype and parenting style. Alternatively, such correlations can occur when a child (perhaps a stubborn child) evokes negative parenting response from a parent who did not (as my wife claims) transmit said stubborness genes – an example of an evocative GxE interaction. In this study, the team examined the mother’s genotype at a 40-bp repeat polymorphism in the 3’UTR of the dopamine transporter (DAT) gene. This is an apt candidate gene, since animal models of DAT loss-of-function show disrupted maternal behavior.
As an initial step, the team evaluated whether maternal genotype was correlated with maternal parenting style. They found that the 10-repeat allele of the DAT gene was associated with more of a negative style of parenting. However, the association of the 10-repeat allele of DAT was rather stronger in mothers whose children were categorized as disruptive than among mothers whose children were categorized as compliant – an example of an interaction of the mother’s genotype with her child’s disruptive behavior (which itself may be due to genes inherited by her – and so on – and so on).
Hard to pin down the genetic blame somewhere here. Maddening actually. Maddening enough to make dealing with my unreconcilable 5-year old seem a simple and welcoming task.
My child carries genetic risk for mental illness. Now what do I do?
Posted in Uncategorized, tagged Anxiety, Development, Genetic testing, Mental disorder, Twin on April 13, 2009| 1 Comment »

- Image by giumaiolini via Flickr
As the personal genomics era dawns, it becomes clear that the new genetic information will lead to more new questions than answers. Consider a well-intentioned parent who finds any number of suspicious risk factors in the genome of their child. Perhaps a genetic risk variant for mental illness – an anxiety disorder perhaps? What can be done? What, if anything, should be done?
Of course there is no simple answer to this question. Nevertheless, the technology itself may create strong demand for answers in the near future. If it were me, I certainly would want to know – something, anything – to help. Furthermore, there are already examples of willful misinformation in the consumer genetic marketplace that seem to prey on anxieties of parents, and which could ultimately heighten the need for reliable, evidence-based guidance.
To this end, the recent research article entitled, “A Genetically Informed Study of the Association Between Childhood Separation Anxiety, Sensitivity to CO2, Panic Disorder, and the Effect of Childhood Parental Loss“[Arch Gen Psychiatry. 2009;66(1):64-71], caught my attention. In this article, the authors consider Panic Disorder, a condition which can lead to the disruption of a healthy personal and professional life. Genetic studies have shown that specific genes can contribute to the risk of the disorder, but also that these genes interact with early life and adult life experience. What might these genes be doing in early life – and if we knew – then might we intervene early on to prevent the onset of the disorder later in life?
Again, there are more questions than answers here, but the research team of Battaglia et al., show – using 712 young adult twins – that a common genetic factor underlies childhood separation anxiety and the adult onset of panic disorder. Thus, it may be the case that the sames genes that contribute to the risk of panic disorder, also may contribute to a form of childhood anxiety. Having found evidence for a particular form of developmental continuity, the research team is one step closer to learning how a genomically-guided child-based early intervention might be structured.
Because there are many pathways that can lead to mental illness and many ways in which the genome interacts with the environment – it will be complex, if not impossible, to design early interventions that prevent the onset of mental illness. In most cases, it is rather likely that most children who carry risk for mental illness, will – due to the probablistic nature of gene-gene and gene-environment interactions – just develop typically and not develop mental illness. Neverthess, some will and its worth learning more.
Genome prepares us for certain environmental cues: “I was expecting that!”
Posted in Visual cortex, tagged Development, evolution, Twin on March 31, 2009| Leave a Comment »

- Image by cobalt123 via Flickr
Is the human brain a blank slate? or a pre-programmed machine that is ready to take the S.A.T.s right out of the box? Obviously neither, or both as it were. Some have gingerly waded into the nature vs. nuture debate and suggested that the human brain comes pre-wired to receive certain experiences – experience expectant – and thus acknowledge the importance of natural selection in shaping an organism via heritable factors but also the need to be able to use the brain to learn from experience and adapt on the fly.
In their paper entitled, “Nature versus Nurture in Ventral Visual Cortex: A Functional Magnetic Resonance Imaging Study of Twins [DOI:10.1523/JNEUROSCI.4001-07.2007] Thad Polk and colleagues provide a wonderful example of this. The team suggested that the brain (visual system) should be somewhat innately (genomically if you will) prepared to process visual stimuli such as faces and objects, but not so for stimuli such as pseudo words. They proposed to test the role of the genome by comparing patterns of brain activity in identical vs. fraternal twins. If the brain activity patterns were very similar for identical twins, and less so for fraternal twins, then it is likely that the genome plays some role in the generation of brain (at least with respect to blood flow) responses to such stimuli. The team used fMRI to assess 13 pairs of identical twins and 11 pairs of fraternal twins for their brain responses to pictures of faces, houses, chairs and non-word strings on letters as well as control “scrambled” images that were comparable in visuo-spatial frequency.
Interestingly, the team found that for faces and houses, there were significant identical vs. fraternal differences in the “activation maps” of the twins but no such differences for chairs and pseudowords. Thus it seems that the genome plays a role in the way the brain processes faces and houses (or perhaps faces and places in general), but not so much for items that are not found (or weren’t found by our evolutionary ancestors) in a natural setting.
I’m surprised by the chair result … although perhaps being a couch potato is something evolution does not select for.
Genes encode white matter pathways to higher intelligence
Posted in White matter, tagged Development, Intelligence, Twin on February 24, 2009| Leave a Comment »

- Image via Wikipedia
Have you ever had your butt kicked by a 12-year old girl? OK, maybe when you were an 8-year old boy perhaps – but I mean as a grown up. Its a humbling experience. I know. For once back in college, I sat for a math contest and was amazed by a young girl who was able to answer each question more quickly and accurately than anyone else (other college students) in the room.
How did she do it? What was different about her brain than mine (illicit substances aside)? Now, as a parent of children who will, themselves, soon start sitting for exams and contests – wouldn’t I like to know. Might I perchance endow my children with brain power? Not likely I imagine, but what is brain power anyway? and what is it about the brain that makes some people perform better in general intelligence assessments? In their new research article, Genetics of Brain Fiber Architecture and Intellectual Performance [doi: 10.1523/jneurosci.4184-08.2009], Paul Thompson’s team of neurobiologists explore this longstanding question.
In this article, the team asks whether the white matter of the brain (the glial cells that ensheath neuronal axons) might be both heritable and correlated with measures of intelligence. To measure white matter integrity, the team uses an imaging method known as diffusion tensor imaging (DTI). It has been shown previously that measures of intelligence are correlated with white matter integrity – presumably because white matter serves as a kind of insulation that speeds up the transmission of action potentials and thus facilitates interhemispheric communication and other long-range forms of neural network processing. The team found that white matter integrity was correlated with performance on intelligence assessments in brain regions such as the cingulum, callosal isthmus, corona radiata, internal capsule and other regions. By imaging 23 pairs of identical and 23 pairs of fraternal twins, the team also found many regions in the brain where white matter integrity was under more than 50% genetic control – particularly in the parietal lobe. Lastly, the team found that in many of these regions, the correlation between white matter integrity and intelligence could be explained by the same genes.
Amazing research findings indeed, that points to where in the brain and what type of physiological processes are related to efficient brain function.
Happy 200th birthday Charles Darwin ! Here’s an inherited acquired characteristic for you
Posted in Hippocampus, tagged Development, Epigenetics, evolution on February 9, 2009| 1 Comment »
Few may pause on February 12 to note the 200 year anniversary of the birth of Charles Darwin and 150 years since the publication of “On the Origin of Species” (click here to download). To some extent, this may be expected since much of the controversy (creator vs. autonomous biochemical processes) seems to have abated – NOT. Politically, the issues are still red hot – in Kansas – and elsewhere across the globe.
But what about the science ? Do we accept the basic tenets of evolution by way of genetic variation and natural selection ? For goodness sake, I mean, we’ve just about (or soon will have) sequenced every living organsim-on-the-planet’s genome. Surely there is no doubt about the validity of the so-called neo-Darwinian synthesis of basic/population genetics and the theory of evolution by natural selection. Is there ? Perhaps you can’t blame folks for trying to poke holes (as covered extensively by Sandwalk), especially on the big 200th anniversary.
One place where I am hearing some buzz on the teetering of neo-Darwinism and the Modern Synthesis lately is in the area of epigenetics. Consider the paper by Arai et al., entitled, “Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment” [doi: 10.1523/jneurosci.5057-08.2009]. In this paper, the researchers measured a trait known as long-term potentiation (LTP), wherein a synapse fires in a longer and stronger fashion. This type of potentiation is thought to be a basic mechanism that neural networks use in learning and memory formation. In their paper, the team found that certain synapses in the hippocampus were potentiated when animals were exposed to an “enriched” environment (normally mice are caged in empty bins lined with woodchips, but an enriched environment is one filled with tunnels, hidden passages, toys, ropes to climb & other stuff to discover and learn about). The team shows that, in response to an enriched environment, the mice acquire the LTP trait.
The next thing the team found was that the offspring of female (but not male) mice that had acquired the LTP trait – did also show the LTP trait – even when they, themselves, did not experience the enriched environment. Thus, the so-called acquired trait (LTP) was inherited by the offspring. Hmmmm – sounds a bit Lamarckian to me, or, as the authors of this research article suggest, “Lamarckian-like”. Is this a case that violates core tenets of the modern synthesis ? Does it besmirch Darwin on his 200th birthday ?
No. Here’s why in a nutshell. The LTP trait is not passaged via the female germ line. That is, the physiological and genetic (gene expression) changes that lead to LTP in the mothers are not encoded in the genome of her eggs. Indeed, her haploid egg cells were set aside long, long before she ever experienced the enriched environment and acquired the LTP trait. Rather the effect is one that seems to be dependent on her uterine environment and ability to transfer information from unterine milieu to developing offspring – whose developing brains seem to be endowed with the molecular components needed to facilitate LTP. Figure 4c of the paper shows that the LTP trait was lost in the F2 generation – therefore the effect is not stably transmitted via the germline (as plain vanilla DNA mutations are).
For an overview of the complexities of incorporating the Central Dogma of Molecular Biology into the Theory of Evolution by Natural Selection, read chapter 4 (p76) Weismann, Lamarck and The Central Dogma of John Maynard Smith‘s book “The Theory of Evolution“. Maynard Smith credits August Weismann’s germ plasm theory as a key factor in the modern synthesis since – by sequestering the germ line very early in development – acquired characteristics cannot be inherited via egg & sperm. Hence, Lamarckian evolution is (in principle) not possible. This seems to be the case here with the LTP trait. In this spirit, the authors do a great job of reviewing other similar examples of how a mother’s uterine environment can lead to epigenetic modifications (click here for review article and here for a PLoS paper on the topic) – such as the viable yellow locus in the mouse [PMID: 18673496] and the effects of endocrine disruptors on methylation of germ cells [PMID: 16973726].
Well, it is amazing indeed how Darwin’s work continues to inspire us. Happy 200th Birthday !
![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)

![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_c.png)

